November 11, 2025
Service Overview
An overview of the Scientific Computing (SciC) Linux Clusters with information relating to the service can be found in the Computational Resources section of our website. The cluster webpage also contains links to the account sign-up form, request forms for applications or storage and the support and feedback forms. Review the typical uses for each of the computational resources offered by ERIS to determine the most suitable platform.
Information about using the cluster is found in the Computational Resources and Analyze Data sections of our KnowledgeBase.
Details of our introductory training courses for Linux and SciC Linux Clusters are in the Scientific Computing section of our Training Page
Obtaining an account
First determine the most suitable platform for your project. Then complete the SciC Linux Clusters account request form, or the corresponding form for the compute platform selected. Estimating the storage, applications and compute time required for your projects will help us provide these services. If you will require access to an existing data folder for your lab / project, find out from the data owner the name of the corresponding group in order to request this access on the account request form.
Once registered your account should be ready in 24-48 hours.
Connecting to the SciC Linux Clusters
Before connecting to the ERIS cluster with any of the methods described below please ensure you have VPN switched on if you are located offsite.
Command-line SSH
Opening a command-line terminal session using an SSH client is the most common way to connect to the SciC Linux Clusters. This method requires some familiarity with the Linux operating system.
The following platform is available to all users following eris cluster registration.
ERISTwo: eristwo.partners.org
and where users submit jobs to the approximately 75 CPU compute nodes using the Slurm job scheduler.
Separately, groups can sign up to access GPU resources on the ERISXdl platform and where subsequently group members can access this node at:
ERISXdl: erisxdl6.research.partners.org
Users submit compute jobs to the 4 x DGX compute nodes (each with 8 x V100 GPUS) using the ERISXdl/Slurm job scheduler.
Connect from a PC running Microsoft Windows:
- (Recommended) Use the MobaXterm application to connect.
For more information on the features of MobaXterm, reference their documentation.
- Download the Free Home Edition of MobaXterm here and extract the files from the download to your preferred location. Run the installer and then application if you downloaded the Installer edition, or run the .exe file if you downloaded the Portable edition.
- Once the application is open, select "Session" from the top toolbar. This will open a new pop-up window.
- Select "SSH" as the session type, and fill in the specified fields as followed. Once done, click "OK"
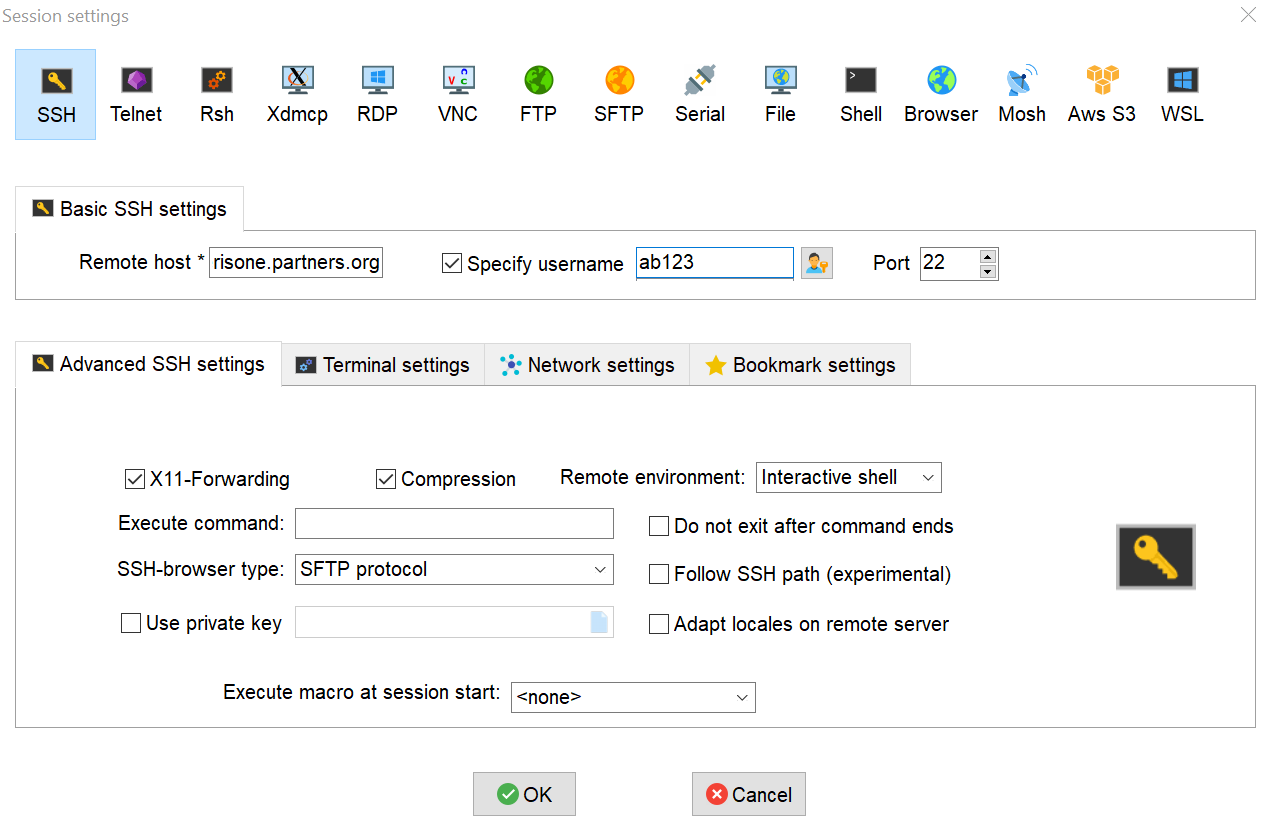
- Remote host: use the host chosen above
- Specify username (make sure it's checked off): your Mass General Brigham user ID
- Port: 22
- A new terminal tab should open in the MobaXterm window, prompting for your password. Enter your Mass General Brigham password to continue.
- Once logged in, you will also be able to view your home directory files and folders in the "Sftp" tab on the left toolbar in MobaXterm
- Use the PuTTY ssh client to connect
- Download the PuTTY ssh client (putty.exe)
- Double click the downloaded file
- Enter the following connection information and press "Open"
- Host Name (or IP address): use the host chosen above
- Port: 22
- Protocol: SSH
- Enter your Mass General Brigham user ID in lowercase and password when prompted
Connect from a Mac or Linux computer
- Open the "Terminal" application
- replace "abc123" with your Mass General Brigham user ID in lowercase and "yourhost" with the host chosen above, and type this line followed by your Mass General Brigham password:
ssh abc123@yourhost.partners.org
The first time you connect to SciC Linux Clusters, a message will appear showing the rsa2 key fingerprint. Select "Yes" to proceed if the fingerprint matches the following (as of 09/2024):
SHA256:AYERiujz9FYfiO0DTxmsuybPL2OVroyMaEkQoC41724 no comment (RSA)Remote Desktop
A remote desktop is ideal for using graphical applications with data stored on the cluster, and for a persistent workspace. On the webpage linked below, you may toggle between available servers.
Web Applications
Web applications allow running jobs, analytics and visualization using only a web-browser. Sections of the quick-start guide below may not apply if you exclusively use to the cluster through a web-browser.
Several labs have web-portals for running analysis pipelines on SciC Linux Clusters. Applications supported by the ERIS Scientific Computing team include
- RStudio
- JupyterHub
Submit a feature request for other web applications.
Transferring Data
Compute nodes, login nodes and remote desktops...
Within the SciC Linux Clusters, computers are referred to as "nodes". Find out which node you are currently working on from its host name, which is shown in the terminal prompt, or from the output of the "hostname" command e.g.
hostname
eris2n5.research.partners.org
Login nodes
In order not to overload the front end and so for the benefit of all cluster users, operations on the eristwo login nodes (eris1n2, eris1n3, eris2n4 and eris2n5) should be restricted to job submission, file editing and small-scale file copying. Please note that users are limited to 30 processes, 1 day of continuous compute time, and 8GB of memory per process on these login nodes. Jobs that go beyond these limits will be terminated without warning. On the ERISXdl login nodes (i.e. erisxdl1, erisxdl2, and erisxdl3) only the compute time is limited.
Users can view the limits with the following:
ulimit -a
Compute nodes
Any computational and data-intensive work takes place on the compute nodes. Time and resources on these nodes is exclusively granted by a request to the the job scheduler to prevent resource conflicts between jobs. You can request a command-line session immediately on a compute node using the interactive queue. To see the list of hosts, use the bhosts command to see which are available.
Running an Application or Job Script (Job Schedulers)
- Submitting Jobs to the Cluster via LSF
- Queues for Job Scheduling and Resource Reservation on ERISOne
- Queues for Job Scheduling and Resource Reservation on ERISTwo
- Getting Started with Slurm on ERISTwo
- ERISXdl Getting Started
To use applications and software provided by the cluster during your jobs, you can take advantage of the cluster's module system. Learn more about loading ready-to-go applications in Loading Applications.
Help and support for SciC Linux Clusters Services
For questions related to the use of SciC Linux Clusters, first check the frequently asked questions in our Knowledge Base. For service request, software installation requests and reporting problems fill the appropriate form from our Forms Library and contact Scientific Computing Support with further queries.
Note that email correspondence from the Scientific Computing support team includes a message header inviting you to check the status of the support requests online. The ticket site http://tickets.partners.org/ does not use your Mass General Brigham ID or password for log-in. Request a new password using your primary email address as the username.
Using SciC Linux Clusters for Research Purposes
If you are applying for a grant and need to reference these resources, you can use the link below. This is to help researchers copy and paste general, high-level, descriptions of infrastructure into their grants, IRB proposals, contracts, etc.
http://rc.partners.org/boilerplates
It would be very much appreciated if the Enterprise Research IS group and/or individual Research Computing members receive acknowledgement in publications if you feel our service and facilities have helped your projects. You can find a guideline for such acknowledgement at:
https://rc.partners.org/research-apps-and-services/boilerplates-templates/it-infrastructure#citing-eris
Please let us know how ERIS helped your research!
https://rc.partners.org/research-apps-services/boilerplates-templates/research-thumbs
