In response to the fast-growing number of COVID-19 research projects, new and enhanced research data tools and data sets are being made available to support these endeavors. To view detailed information about these tools and review data within each, please visit Digital Research Dashboards and data dictionaries hosted by Collibra. The goal is to support the rapid turnover of analytics to help researchers understand the signs and symptoms, comorbidities, and treatments of COVID-19. Researchers can leverage any combination of these tools to support COVID-19 research.
- COVID-19 Data Mart housed in the Enclave
- COVID-19 Detailed Data Files housed in the Enclave
- COVID-19 Vaccine Registry housed in the Enclave
- COVID-19 Summary Table
- COVID-19 External Data Sets
- DICOM Images for Chest X-Ray with eUnity Viewer
- RPDR (Research Patient Data Registry) COVID-19 patient data updated daily
- Biobank Portal with COVID-19 defined patients with genomic information
- COVID Biobank Portal
COVID-19 Research support email: MGBAnalyticsEnclaveSupport@mgb.org
The Enclave allows the analysis of multiple types of healthcare data in a secure environment.
The COVID-19 Detailed Data Files contain derived data from the COVID-19 Mart, which was denormalized and transformed for easy consumption.
The COVID-19 detailed data files are flat files that are derived from the COVID-19 Mart tables and provide a quick and clear delineation of specific data without complex querying of multiple tables or a large database. The data was denormalized and pre-processed to facilitate data input for machine learning tools and data science analysis. The COVID-19 detailed data files reside in a high-security Enclave platform.
Current categories of data in the files include Demographics, Diagnosis, Labs, Medications, Procedures, Vitals, Encounter, Providers, RPDR concept, and Patient MRN. These files are derived from the related tables in Analytics and Dimension schemas in the COVID-19 Mart.
Detailed Data Files
- The data is available in flat files format and is static up to August 2023. If the latest data files are needed, researchers should build a data request in the RPDR.
- The files are tab-delimited, which is an easy-to-read format that can be opened with R, Python or SAS.
- A data dictionary describing each table’s data values can be found in the same location as the detailed data flat files.
- Review the available in the same directory ReadMe file for additional information.
How to Access
Access to the COVID-19 detailed data files is dependent on provisioned access to the COVID-19 data mart housed in the Enclave. Before requesting access to the COVID-19 Mart (and consequently the Detailed Data Files), please review the Digital Research Dashboard: COVID-19 Mart in Enclave, as well as the requirements for requesting access. When ready, please follow the link to Enter your access request. Once your access request is submitted, the turnaround for granting access is 1-7 business days. If you are a Project Staff Member, your Project Leader will be requested to approve your access.
Questions
- For information or questions regarding the detailed data files, or to learn more about the Analytic Enclave's platform and tools, contact the COVID-19 Mart support team at MGBAnalyticsEnclaveSupport@mgb.org.
The registry includes identified patient vaccination data for patients with a record of receiving any type or dose of COVID-19 vaccine.
The COVID-19 Summary Table is a spreadsheet that contains summary information on the defined COVID-19 positive patient cohort
The COVID-19 Summary Table contains discrete data elements for quick identification and analysis of the MGB COVID-19 positive patient cohort. These include patient demographics, EPIC Infection flags, COVID-19 PCR, and antibody laboratory tests, inpatient admission infromation and phenotype data.
Research opportunities for using this data include:
- Evaluating the trajectory of infection by analyzing the first identification of COVID-19 (by EPIC infection flag or PCR lab test) to the most recent negative antibody laboratory test.
- Determining clusters of infection based on demographic location, age, or gender.
- Calculating the mortality rate of the cohort.
- Understanding disparities based on race or ethnicity.
The COVID-19 Summary Table data originates from the MGB COVID-19 Mart found in the Enclave which contains vast amounts of patient data originating from the RPDR and EDW.
How to Access
Before applying for access, please review the Digital Research Dashboard: COVID-19 Summary Table, as well as the requirements for requesting access. When ready, please follow the link to Enter your access request. Once your access request is submitted, the turnaround for granting access is 1-2 business days. If you are a Project Staff Member, your Project Leader will be requested to approve your access.
If you have any questions, please contact MGBAnalyticsEnclaveSupport@mgb.org.
Collection of COVID-19 data available to researchers owned and maintained by other MGB departments
External data sets are stored in either the Data Enclave or Shared File Areas.
Available in COVID-19 Data Mart housed in Data Enclave
Pulmonary X-Ray Severity (PXS) Score developed by the Center for Clinical Data Science
The PXS data consists of a numerical PXS score for over 8,600 COVID-tested patients. It is accessed directly in the COVID-19 Data Mart within the Radiology schema. Current users will automatically see this data in the Radiology.PulmonarySeverity table starting the Aug 12, 2021 data refresh.
Available in Shared File Area (SFA)
MGH COVID Registry validated by MGH Dept of Medicine. The MGH COVID Registry is only available to MGH researchers at this time
The MGH COVID-19 Registry contains manually reviewed data of over 500 data elements for 1200+ COVID-19 positive patients seen at MGH. Access must be requested separately to view and download these SAS files.
Have data to share? Email MGBAnalyticsEnclaveSupport@mgb.org and attach a filled-out Data Sharing: About the Data template.
Access raw DICOM images for over 19K COVID-19 tested patients and view images.
New as of March 3, 2021
From March through August, Chest X-Rays from COVID tested patients at MGH were run through a deep learning algorithm to calculate a Pulmonary X-Ray Severity (PXS) Score.
The Chest X-Ray images (in DICOM format) are now available to all researchers to conduct their own analysis or review. Images can be easily viewed by looking up specific patients and accession numbers using the eUnity Viewer configured for access to them. The images can also be downloaded for additional analyses, including creation of other algorithms, following MGB guidelines for securing identifiable patient data.
Not only are the PA (posterioranterior) & AP (anteriorposterior) views available that were used in the PXS score generation (70,000 images), but additional images from the exam are also included. There are approximately 125,000 images available in total. The studies are from CXR from MGH & BWH; however, the PXS scores are only available for MGH at this time.
Acknowledgement: Access to the images and eUnity Viewer is provided in partnership with CCDS and EMI.
How to Access
Let us know you want access! Access is currently handled by individual request during this pilot phase. Access permits download of the DICOM files and the specally configured eUnity Viewer, an application from Mach7 Technologies.
To request this additional access, please email: MGBAnalyticsEnclaveSupport@mgb.org
Note: As a pre-requisite, a user must also have access to the COVID-19 Data Mart. If you do not yet have access to the COVID-19 Data mart, please follow this link to Enter your access request.
The RPDR Daily Query Tool is used to identify COVID-19 patients using laboratory tests updated DAILY.
The RPDR Daily Query Tool includes the following COVID-19 laboratory tests updated DAILY. Patients tested for COVID-19 from the previous day can be identified and data requested.
What Data is Available?
In the Query Tool hierarchy of terms, here is what is available under Laboratory tests\COVID-19:
- SARS coronavirus RNA/PCR lab tests;
- COVID-19 test comments;
- COVID-19 lab order result statuses;
- COVID-19 IgG and IgM antibody tests; and
- COVID-19 specimen sources.
Also available in the hierarchy of terms under Infection Control Flags\COVID-19 Flags:
- CoV-Exposed (INFG:67) – manually added/resolved to a patient’s chart by infection control staff
- CoV-Positive (INFG:65) – manually added/resolved to a patient’s chart by infection control staff
- CoV-Presumed (INFG:69) – manually added/resolved to a patient’s chart by infection control staff, assigned when a patient has recovered from symptoms that match COVID-19, but currently have a negative test. Those patients may have been positive, but were not tested within the timeframe that they were presumed to have been positive.
- CoV-Risk (INFG:66) – automatically added via a silent BPA based on the ordering of an outpatient Normal status COVID-19 lab order or outpatient COVID referral order, or automatically based on inpatient ordering/outpatient future or standing ordering of a COVID-19 lab order, automatically based on inpatient ordering of a Risk for COVID-19 nursing order, manually from infection control staff if patient is at-risk
- CoV-Clearance (INFG:68) – has been retired as of 5/11/20, was assigned to asymptomatic patients at low-risk for COVID-19 admitted to the hospital
Researchers can query for COVID-19 patients and request identified data.
Researchers may also navigate to the Previous Queries tab, click "Shared Queries" and run the query for "[RPDR] COVID Notes Search" to obtain COVID-19 related information from patient notes.
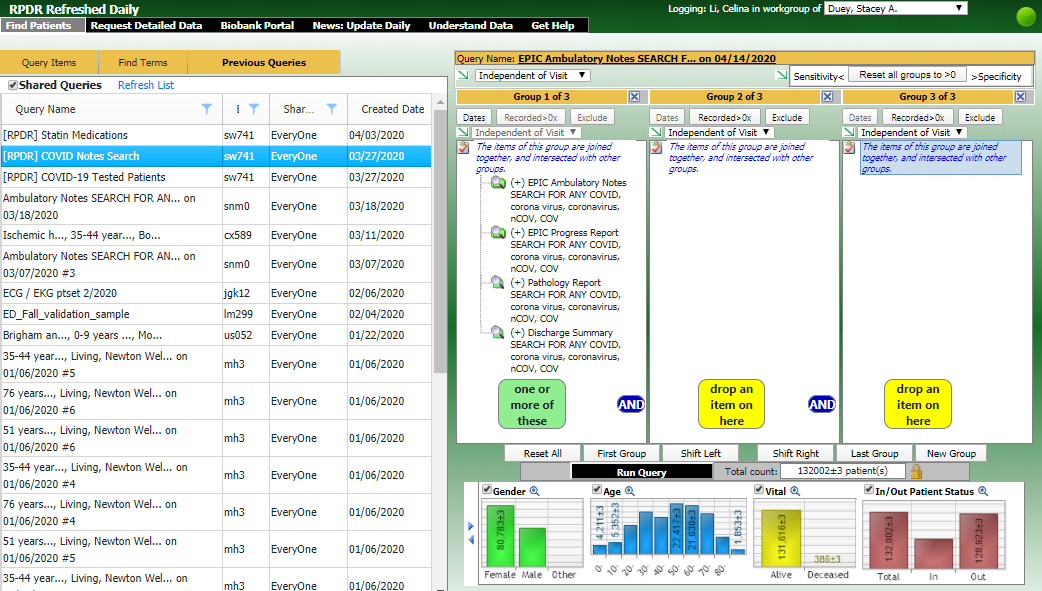
Match Control Functionality
The RPDR Daily Query Tool now allows researchers to identify a control population based on an existing patient population. After researchers have built their query using the Daily Query Tool and obtained an aggregate patient count, they will go to the Match Control tab to obtain an aggregate count for their new control population. When 'Use exact matches only' is selected in the right side panel, it will apply an 'AND' logic where all the selected items in that section must be true in the control population. On the other hand, when 'Use exact matches only' is NOT selected, it will apply an 'OR' logic where at least one item that is selected must be true in the control population.
After researchers click 'Submit request to find set of Control patients,' they will see a new Matched set query appear under the Previous Queries tab.
Once the Matched set query is ready, researchers may select the Matched set query, go to the Request Detailed Data tab and click 'Using a query' to request identified patient information on their newly established control population.
How to Access the Tool:
Additional Resources
Instructions on how to query for COVID-19 patients in the RPDR Daily query tool can be found in the COVID How To document.
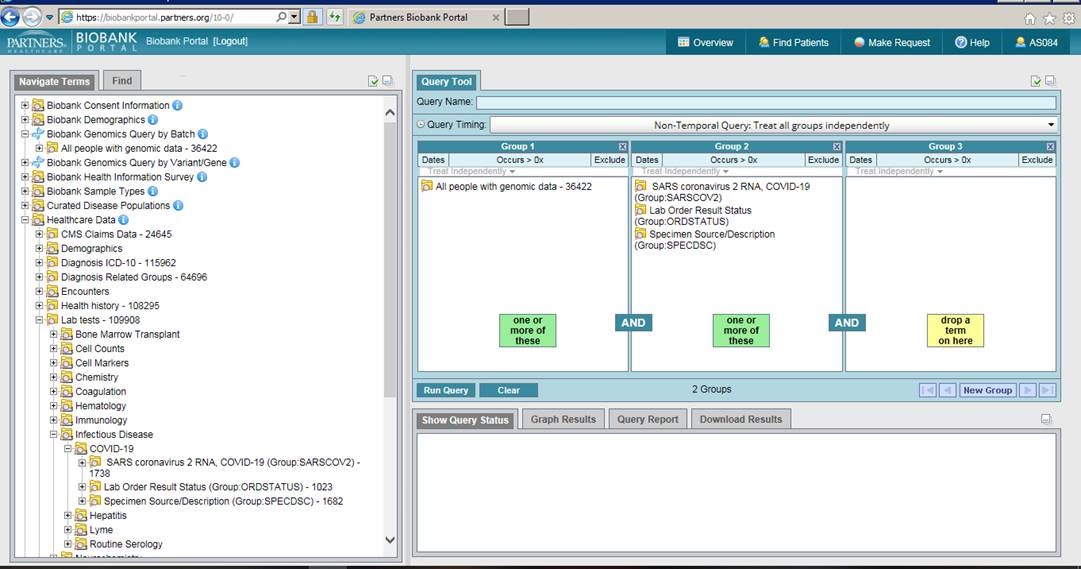
The Biobank Portal is a web-based query tool that allows Mass General Brigham investigators to query and download data about consented Biobank subjects.
The Biobank Portal is a web-based query tool that allows Mass General Brigham investigators to query and download data about consented Biobank subjects. You can also make Biobank sample requests for plasma, serum, DNA, and genomics data directly from the portal.
What Data is Available
Investigators can define a COVID-19 cohort of Biobank consented patients, access pre-disease samples, gather genomic data on a subset of patients who have been genotyped and request identified patient information (structured and unstructured).
To find patients who have tested positive for COVID-19, navigate to the lab tests folder under Healthcare Data and then to the Infectious Disease folder, and then to COVID-19. Drag over the SARS coronavirus 2 RNA folder and select all values of ‘detected’, ‘positive’ or ‘presumed positive’. If you are interested in those who have genomic data, drag the ‘All people with genomic data’ folder into a second panel and run the query. A genomic data request can then be made by selecting the Make Request tab and completing the Request Genomics form.
How to Access the Tool
For more information visit biobankportal.partners.org or email @email.
Additional Resources
The COVID Biobank Portal is a web-based application built to help researchers create custom analytics for observational studies.
The COVID Biobank Portal is a web-based application built to help researchers create custom analytics for observational studies. The underlying population is all subjects who have been consented to the MGB Biobank and have received a diagnostic test for COVID-19 or have a COVID-positive infection control flag. The COVID Biobank Portal enables the creation and download of limited data sets (LDS) and R-generated result tables.
COVID Biobank Portal vs Biobank Portal is illustrated below.
To access the COVID Biobank Portal, go to: https://biobankportal.partners.org/covid/
Additional information and resources can be found on the COVID Biobank Portal Wiki or email @email.
